Examples¶
Co-ocurrence Network Case Study: Clavibacter¶
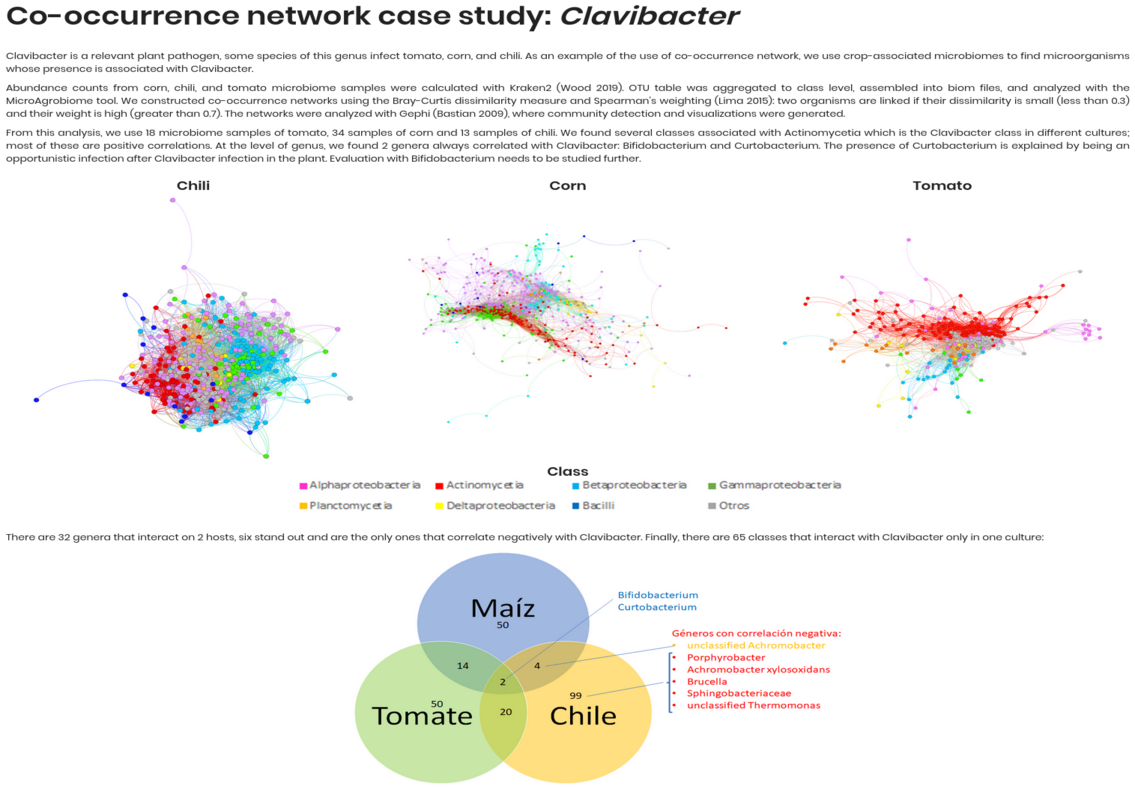
This section provides an example of how to analyze co-occurrence networks using the Clavibacter dataset. The dataset includes various Clavibacter species, and the analysis focuses on identifying microbial genera associated with Clavibacter across plant microbiomes (chili, corn, tomato).
Input:
OTU tables from microbiome samples (Kraken2) aggregated at genus level.
Analysis:
Bray-Curtis dissimilarity and Spearman correlation.
Links defined by dissimilarity < 0.3 and weight > 0.7.
Networks built and visualized in Gephi, colored by class.
Samples:
Chili (n = 13)
Corn (n = 34)
Tomato (n = 18)
Findings:
Bifidobacterium and Curtobacterium consistently correlated with Clavibacter.
6 genera negatively correlated across two hosts.
65 taxa showed host-specific associations.
Includes co-occurrence graphs and Venn diagram summarizing genus overlap across hosts.