Microbiome¶
After submitting your query for Metagenome in the search section of the MicroAgroBiome portal, you will be directed to the Microbiome tab. This section provides detailed information about the microbiome data associated with your selected plant and sample type.
The results will display two tables:
Samples¶
Contains sample metadata such as SAMPLE, HOST NAME, PLANT PART, ASSAY_TYPE, etc.
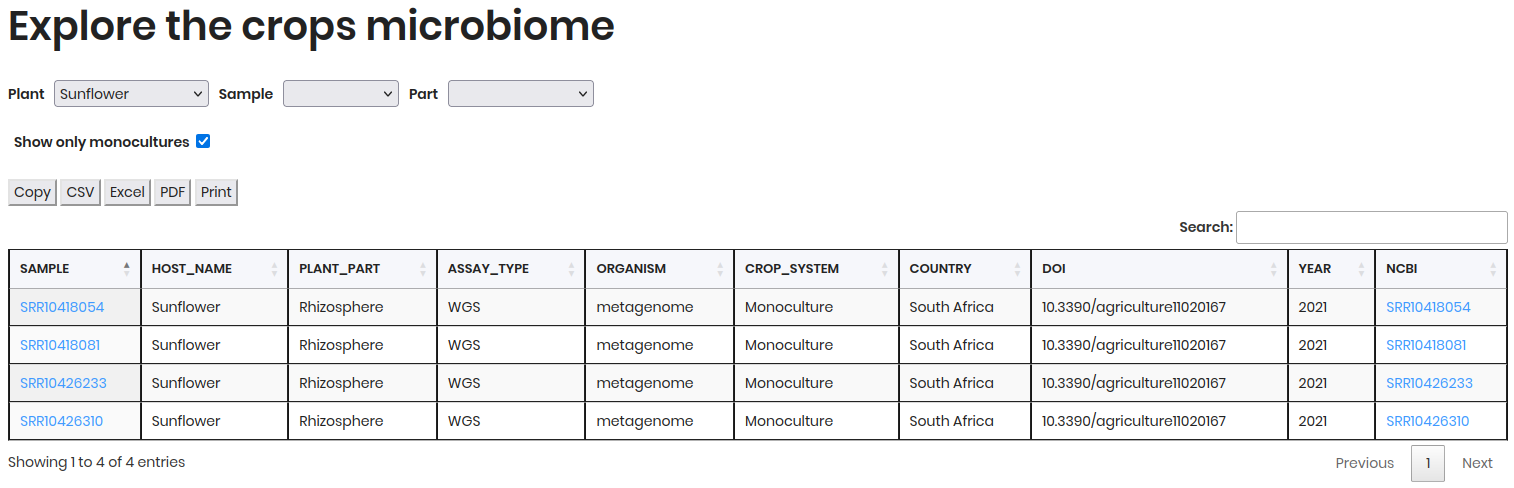
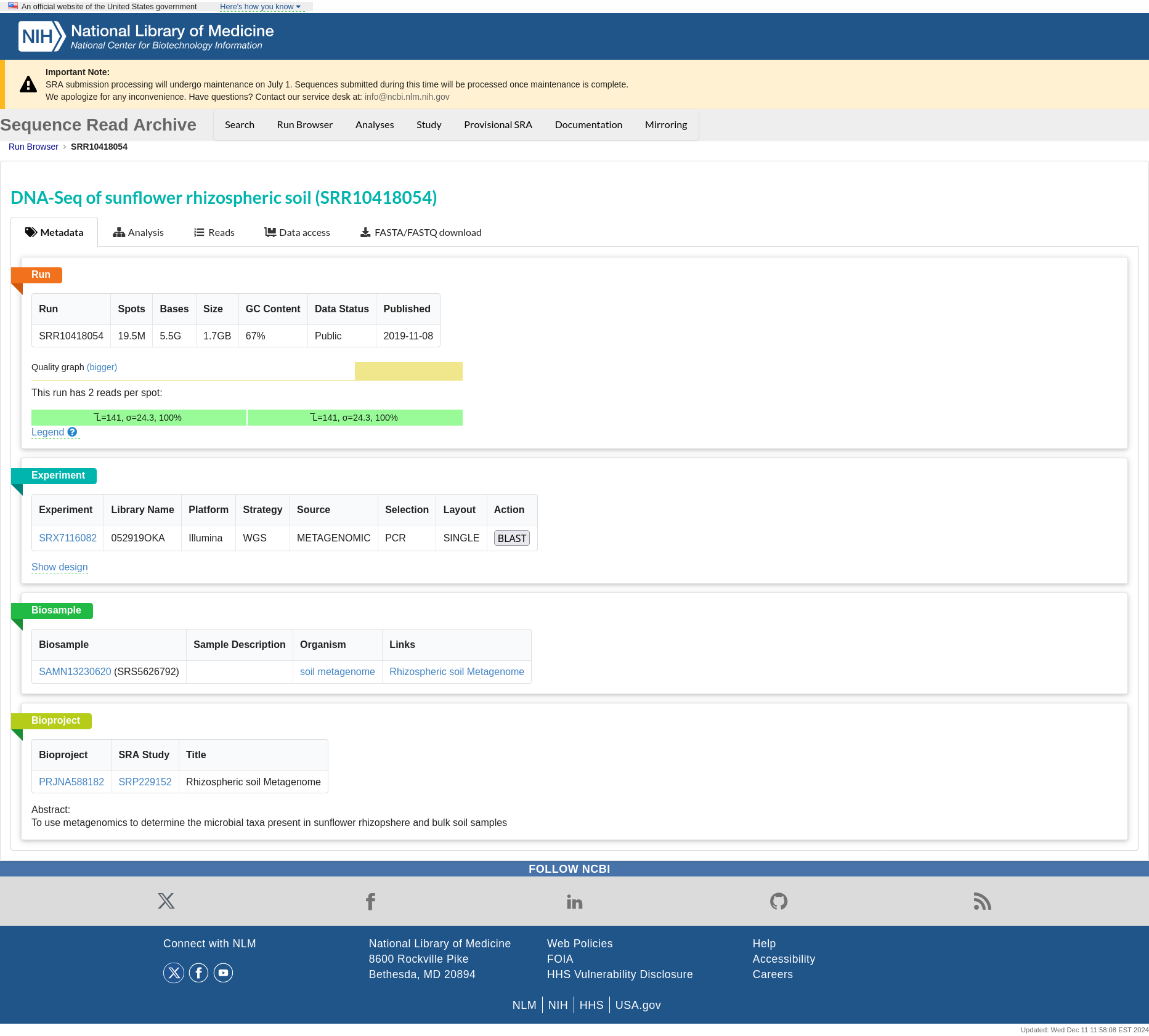
Click on a NCBI to access detailed information about the sample in Sequence Read Archive (SRA).
Click on a SAMPLE to access detailed information about the sample. This information is a Krona visualization of the microbial community composition, showing the relative abundance of different taxa in the sample.
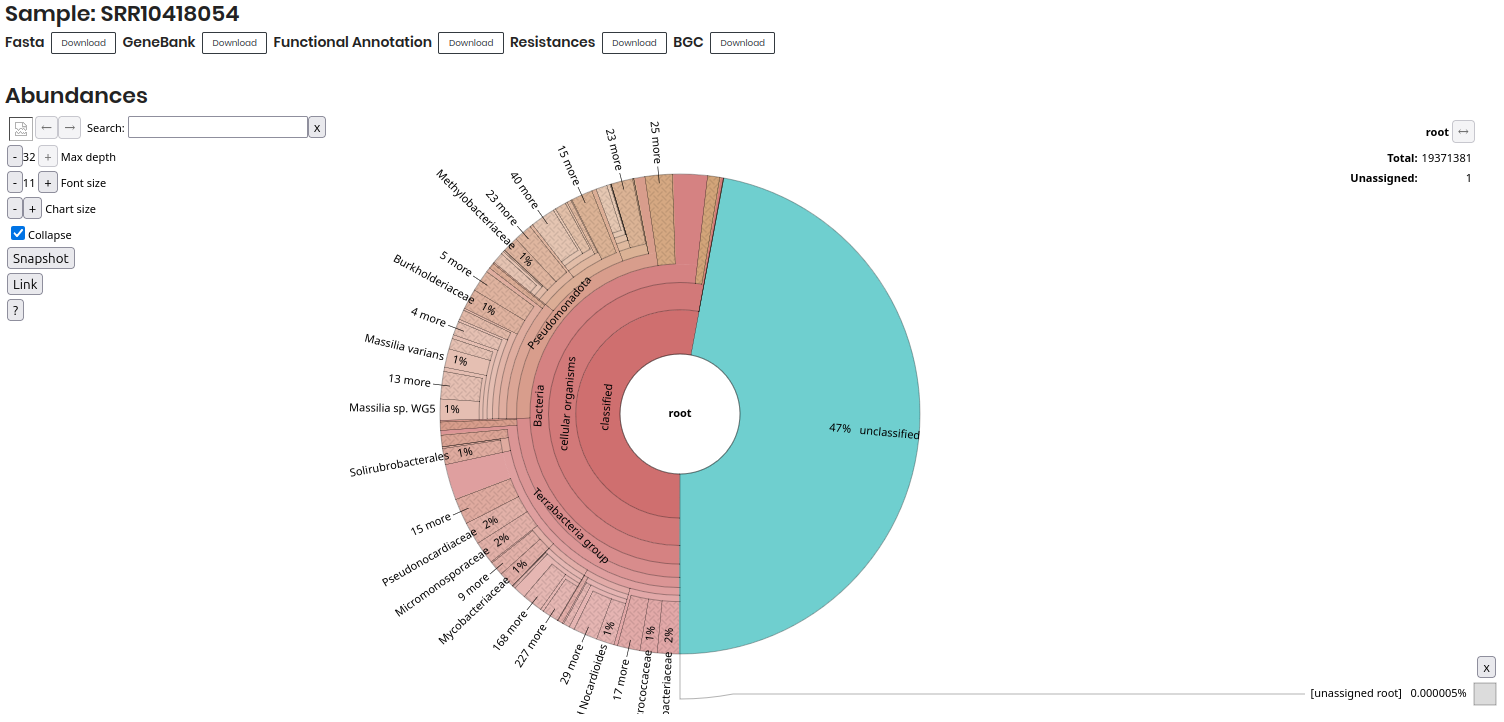
Additionally, you can dowload the different files associated with the sample, such as FASTA, GeneBank, Functional Annotation, Resistances, and BGC files, which are essential for further analysis.
Comparative table¶
Contains detailed data on microbial presence and abundance, including NAME, RANK, TID, MAX, RELATIVE ABUNDANCE of each sample, etc.
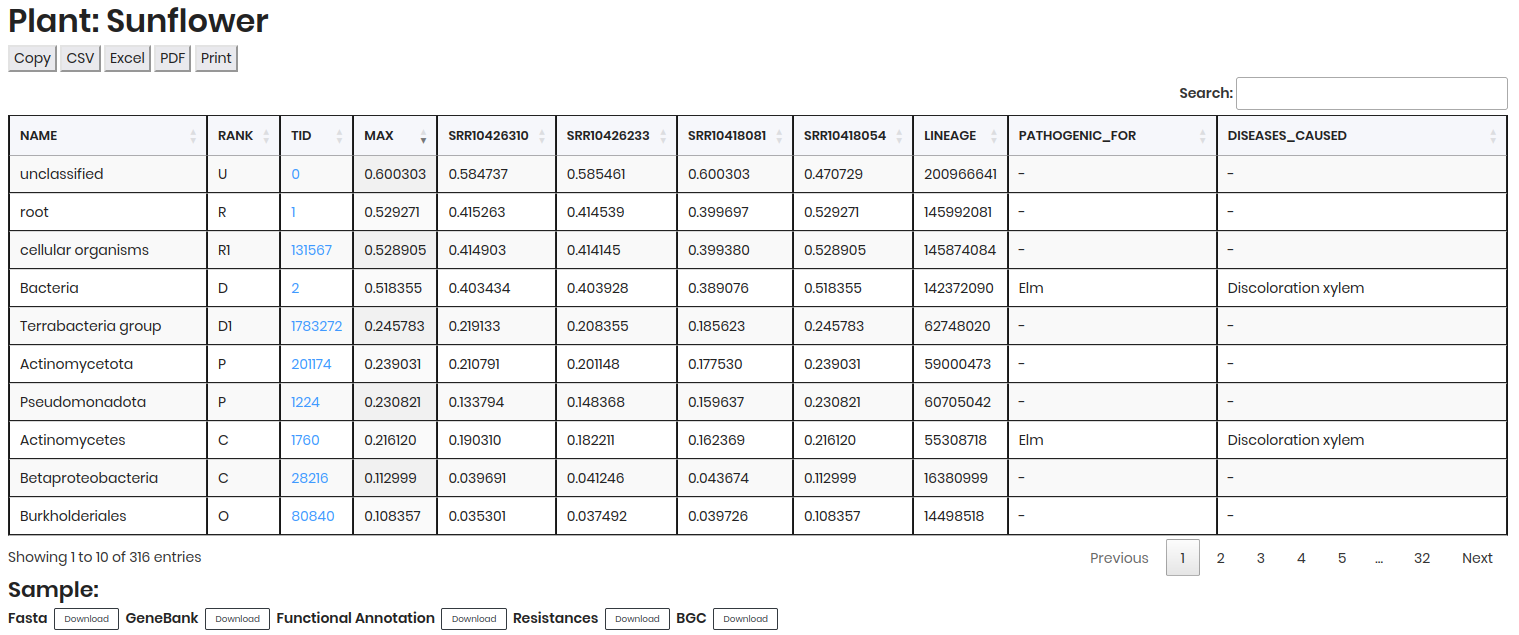
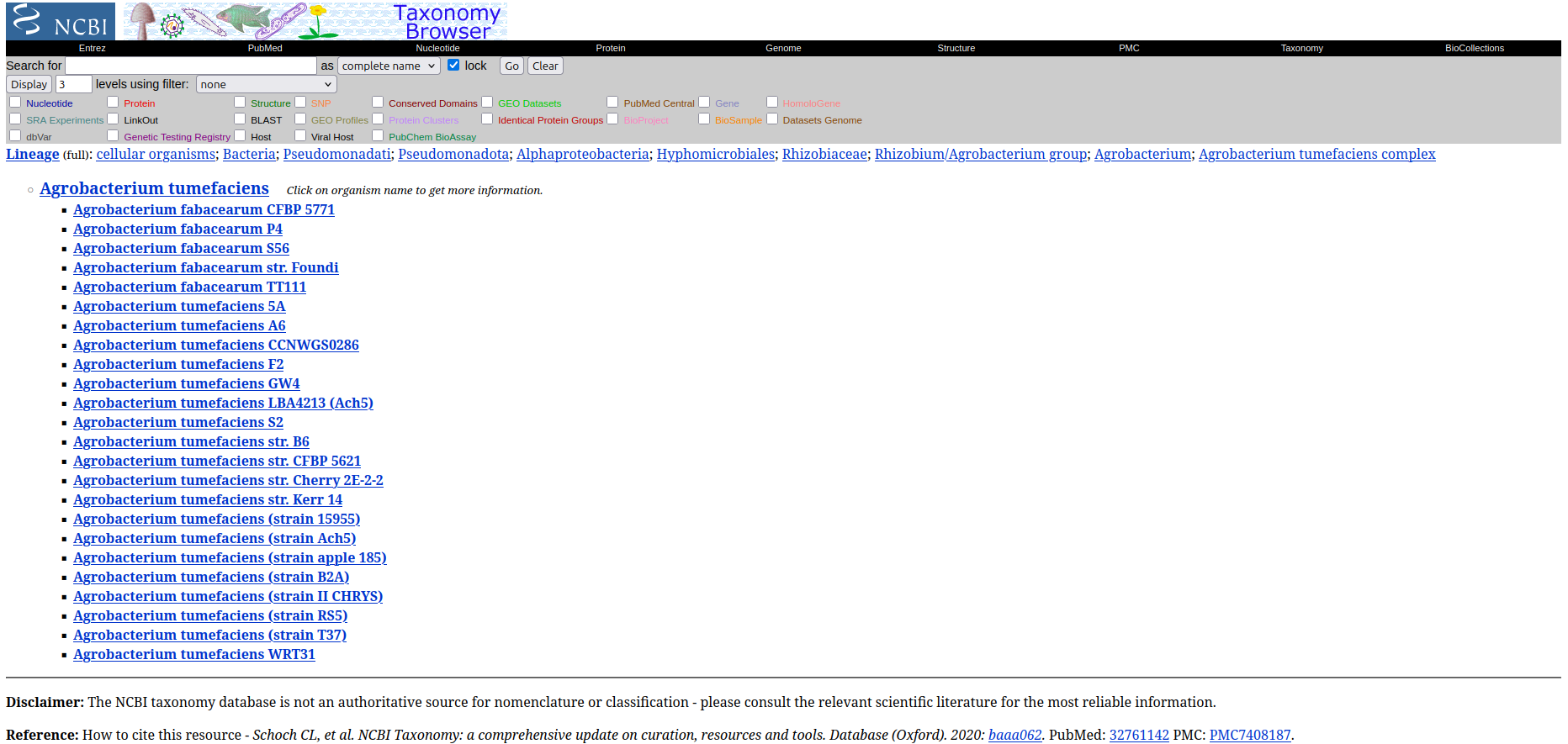
Click in a TID to access detailed information about the taxon in NCBI Taxonomy.