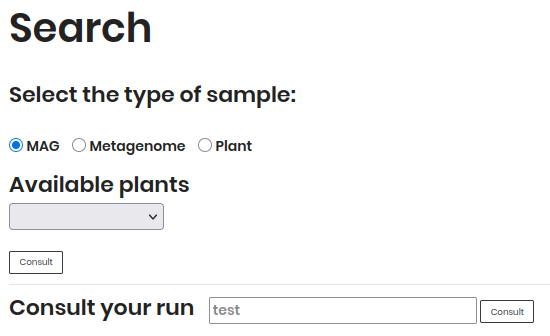
Search Section¶
Filter your search using the following options:
1. Sample Type
MAG (Metagenome-Assembled Genomes): Genomes reconstructed from metagenomic data.
Metagenome: Results of metagenomic samples.
Plant Disease: Data related to pathogenic plant-microbe interactions.
2. Available Plants
Select a plant from the dropdown menu after choosing a sample type.
3. Run Query
Enter a Run ID directly to retrieve your results.
Click Consult to execute your search.
Search Examples¶
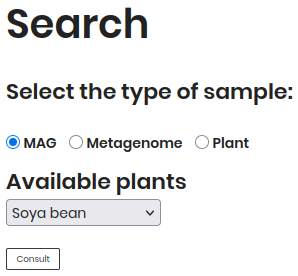
By MAG¶
Follow these steps to explore Metagenome-Assembled Genomes (MAGs):
Select “MAG” as the sample type.
Choose a plant (e.g., Soya bean) from the dropdown.
Click Consult to view results.
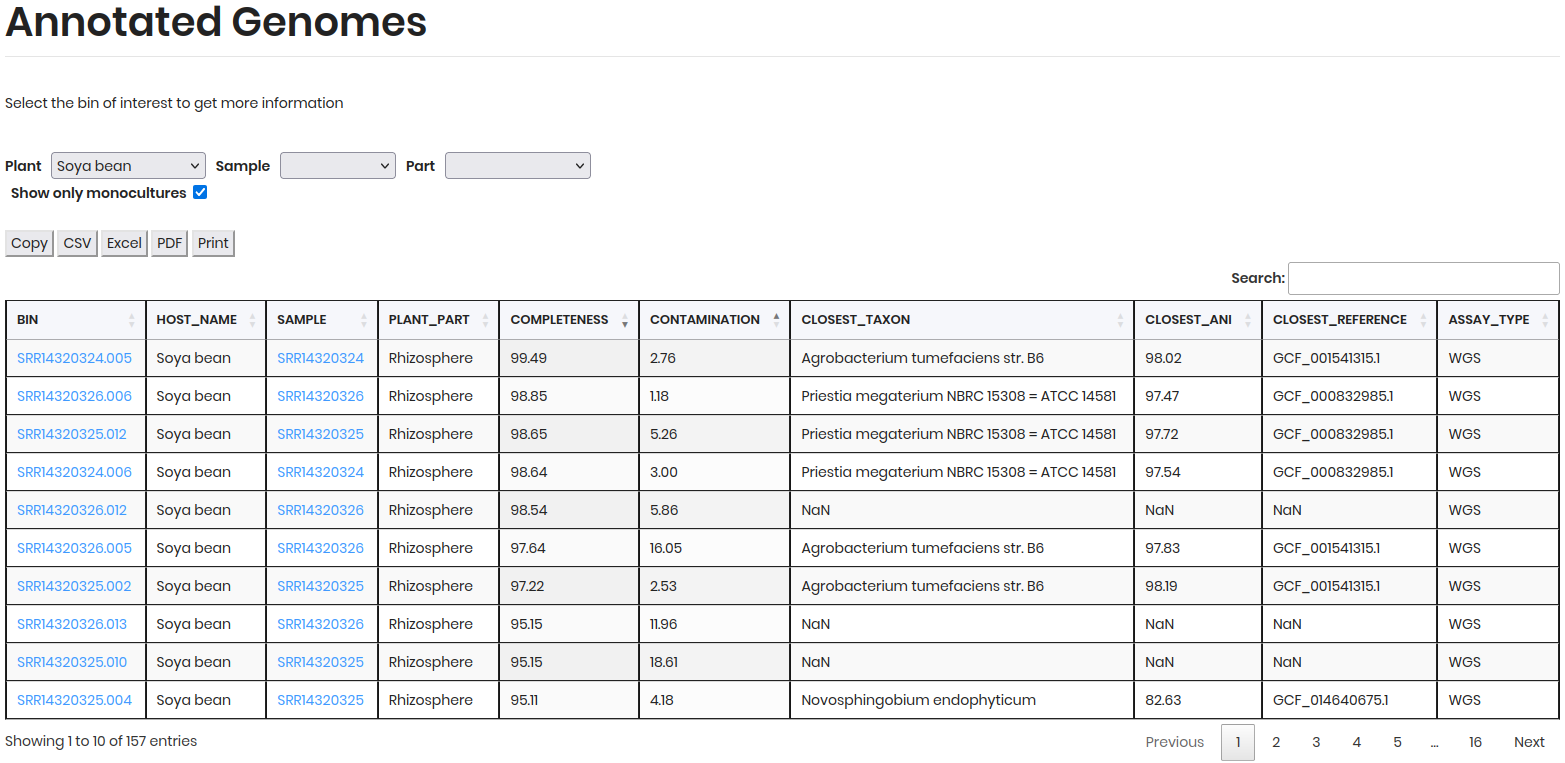
The results will display a table of MAGs, including details such as BIN ID, HOST NAME, SAMPLE, PLANT PART, COMPLETENESS, CONTAMINATION, etc.
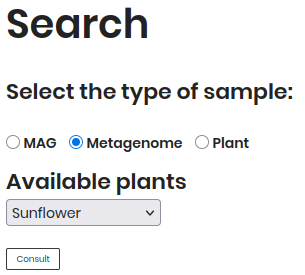
By Metagenome¶
Follow these steps to explore Metagenome:
Select “Metagenome” as the sample type.
Choose a plant (e.g., Sunflower) from the dropdown.
Click Consult to view results.
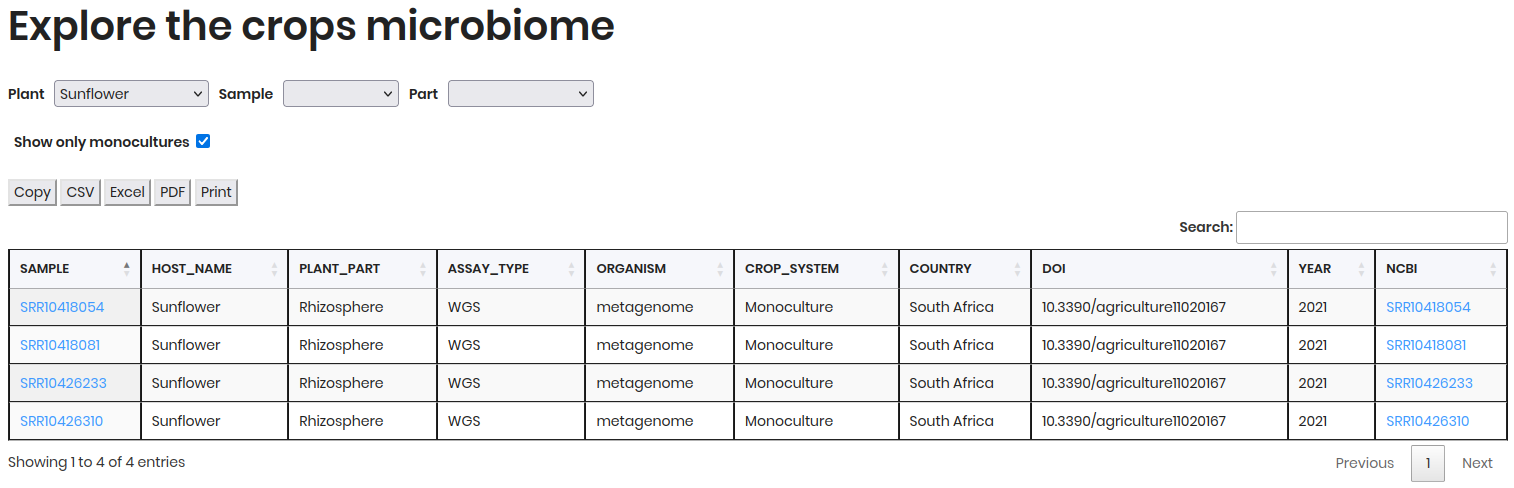
The results will display two tables:
Samples: Contains sample metadata such as
SAMPLE,HOST NAME,PLANT PART,ASSAY_TYPE, etc.
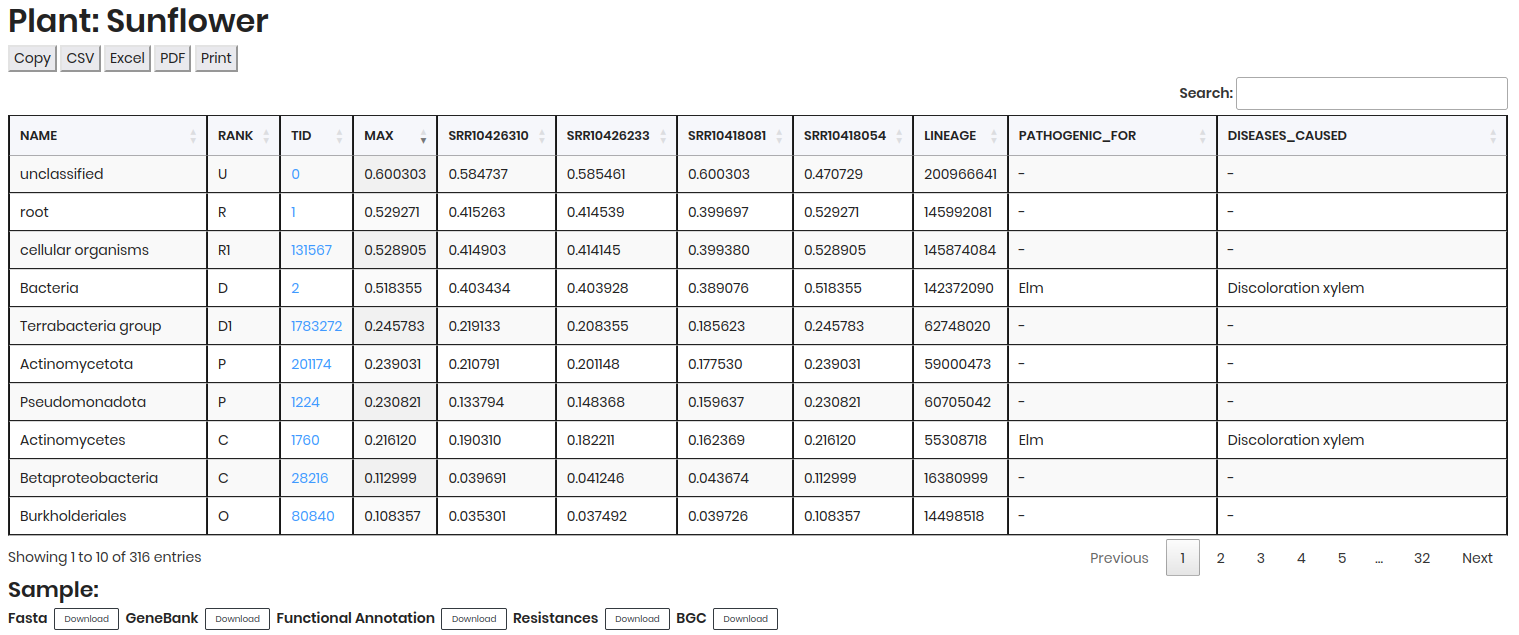
Comparative table: Contains detailed data on microbial presence and abundance, including
NAME,RANK,TID,MAX,RELATIVE ABUNDANCEof each sample, etc.
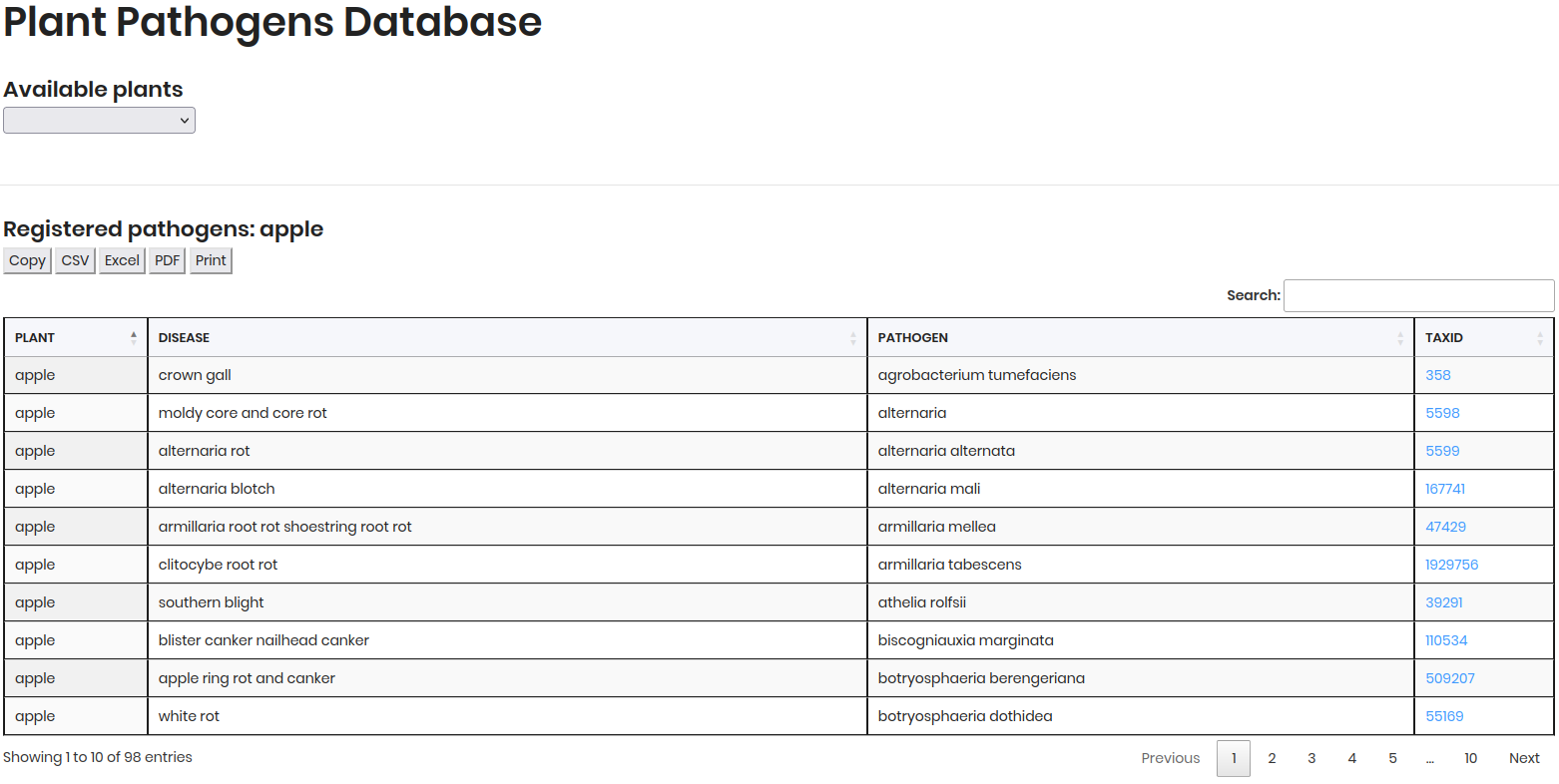
By Plant Disease¶
Follow these steps to explore Plant Disease data:
Select “Plant Disease” as the sample type.
Choose a plant (e.g., Apple) from the dropdown.
Click Consult to view results.
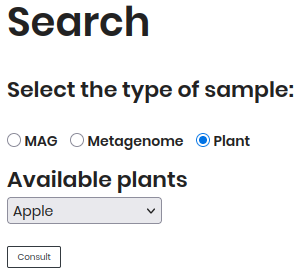
The results will display a table of plant disease data, including PLANT, DISEASE, PATHOGEN and TAXID.